We offer comprehensive analysis of sequencing data for a variety of genomics and other sequencing-based experiments.
We accept most types of sequencing data, including Illumina, Oxford Nanopore, PacBio, 454, and Sanger reads, and have experience with a variety of sequencing protocols (RNA-seq, ChIP-seq, Methyl-seq, whole-genome sequencing, whole-exome sequencing, etc). Using the very latest software, we work with you to design custom algorithms and analyses to fit your study needs.




Examples of Analysis:
Transcriptomics (bulk and single cell RNA-seq)
ChIP-seq and ATAC-seq analysis
Methylation – seq
SNP and structural variant detection
16S rRNA analysis
Custom (includes consultation)
Prices for individual projects will depend on the size and demands of the project, including the need for designing custom solutions. While we cannot estimate the needs for each type of analysis.
Please contact us for a more specific solution and pricing for your project. If you are preparing a grant proposal, please allow at least one week for a custom quote and to discuss your specific needs and analyses.
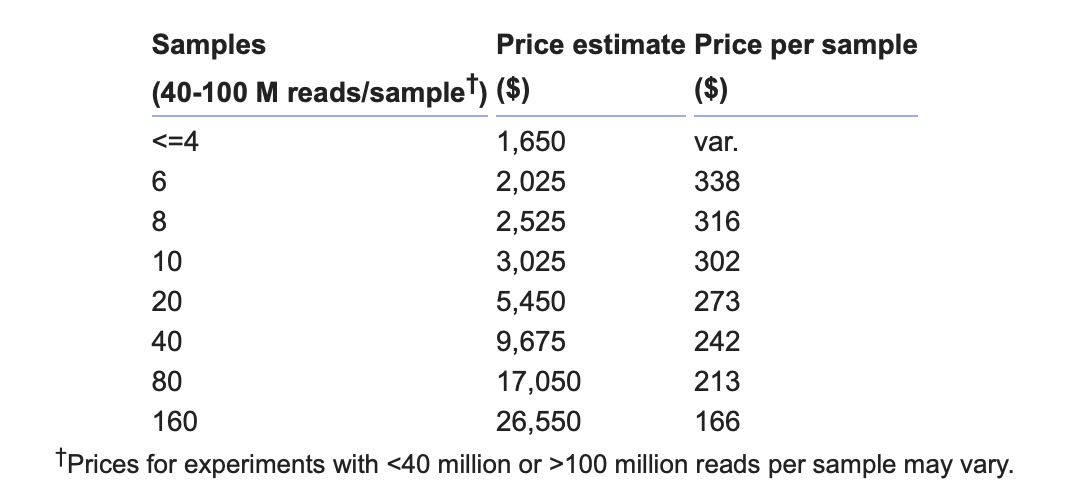
Prices apply to basic RNA-seq analyses with 40-100 million reads per sample. A basic RNA-seq analysis involves read mapping and quality control, transcript assembly and quantification, and computation of differential expression for all genes.
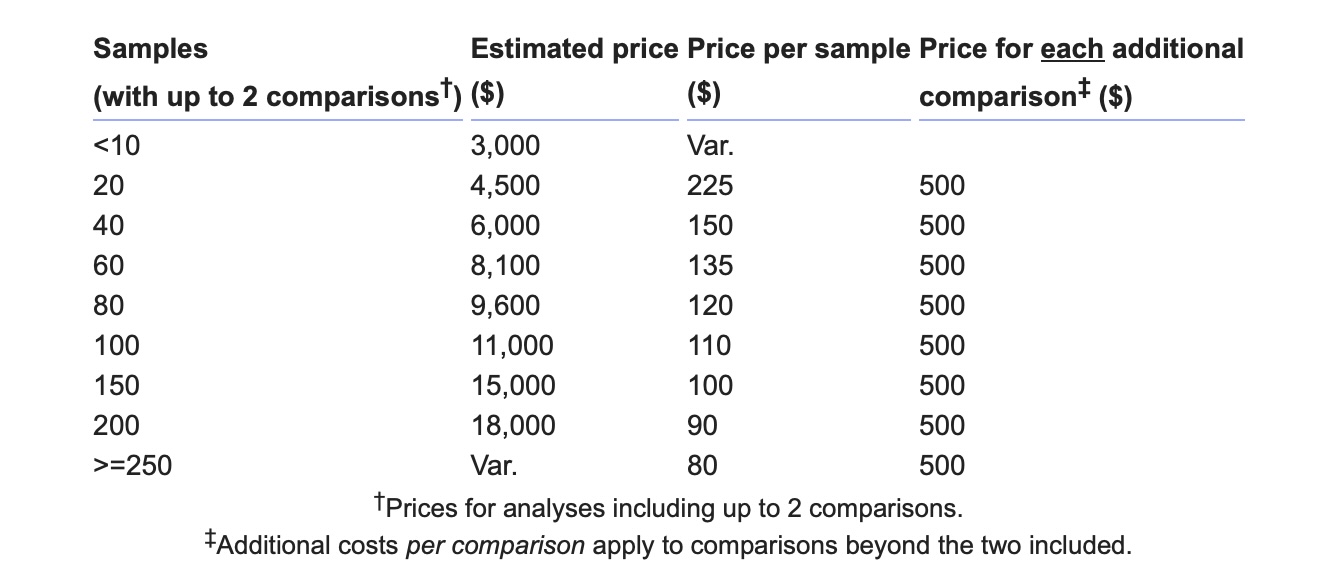
Prices apply to basic 16S rRNA analyses with up to two comparisons. A basic 16S rRNA analysis includes read quality control, OTU assignment, calculation of α and β (PCA) diversity values, and differential taxa analysis, using the QIIME2 and MetaStats packages with the GreenGenes database. Additional costs apply to changes in protocol, and for each additional comparison. For example, an analysis to determine differential taxa in a group of patients before and after treatment includes one comparison. An analysis of patient data before and after treatment, differentiated by the type of treatment, includes two comparisons. An analysis of patient data before and after treatment, separately by type of treatment, and differentiated by gender, entails three comparisons.
Pricing

How do I get started?
To request a consultation follow these steps:
1.Click on the link above
2.Under the service listing select
Computational BioAnalysis
3. Click on
Initiate Request
Contact us
For more information or to submit a request for analytical services please contact:
Liliana Florea, Ph.D.
Associate Professor

